Chromeleon™ 7.2 Chromatography Data System (CDS) Software
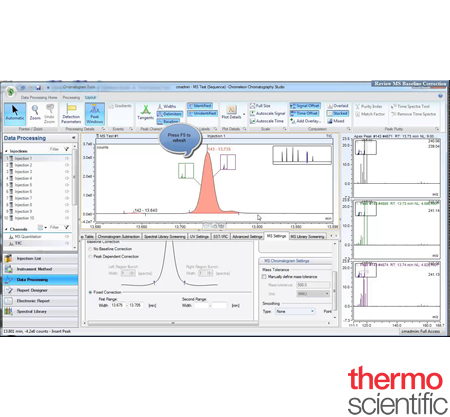
Streamline your laboratory workflow using Thermo Scientific™ Chromeleon™ 7.2 Chromatography Data System (CDS) software. This software delivers superior instrument control, automation, data processing, and more. The first CDS to unify the workflows for chromatography and routine quantitative mass spectrometry (MS) analysis, Chromeleon software provides full integration of Thermo Scientific gas chromatography (GC-MS/MS) and liquid chromatography (LC-MS/MS) instruments. Run your analyses in an enterprise environment—from method creation to final reporting
Enhance Your Workflow Using the Innovative Capabilities of Chromeleon 7 CDS software
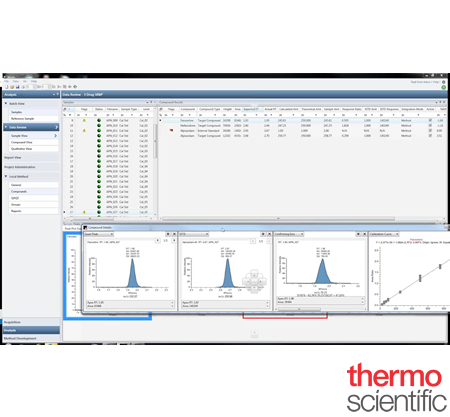
TraceFinder™
Offer increased flexibility and an array of capabilities in performing targeted screening and routine quantitation with either high resolution accurate mass (HRAM) and/or triple stage quadrupole (TSQ) mass spectrometers with Thermo Scientific™ TraceFinder™ Software. Not only does TraceFinder provide method development tools for all molecule types, it also generates new methods from existing data. From pharma/biopharma to food safety, from environmental safety to clinical research and forensic technology, TraceFinder ensures increased productivity for your analytical laboratory, enabling you to address analytical challenges today and tomorrow
Discover the Unique Benefits of TraceFinder
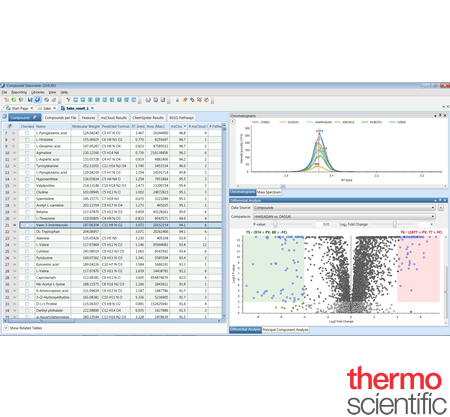
Compound Discoverer™
Streamline and customize data analysis with Thermo Scientific™ Compound Discoverer™ small molecule identification software. Compound Discoverer empowers researchers to strategically collect, organize, store and report data for both targeted and untargeted high resolution analyses. The software simplifies data processing with its ability to customize workflows, assemble data collected from multiple samples into one unified report, and integrate compound identification capabilities. Compound Discoverer also offers a suite of tools for metabolism, unknown metabolomics, impurities and degradants, extractables and leachables, environmental and food safety research, and forensic toxicology
Extensive Toolset
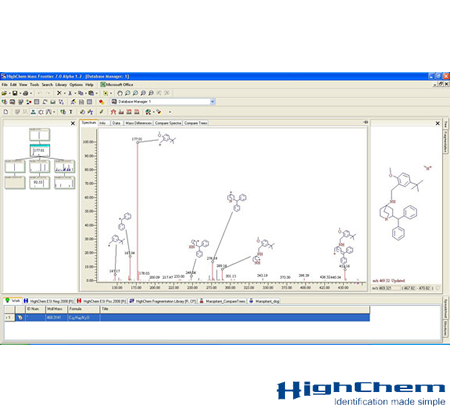
Mass Frontier™
Transform mass spectral data into answers, quickly and easily using Thermo Scientific™ Mass Frontier™ spectral interpretation software. Mass Frontier software provides small-molecule structural elucidation for research into metabolism, metabolomics, forensics, natural products, impurities, and degradants. Innovative features simplify the management, evaluation, and interpretation of GC and LC mass spectral data. Mass Frontier software complements Thermo Scientific™ Compound Discoverer software for small molecule research applications
Intelligent & Easy to Use
ToxFinder™
Clinical researchers and forensic toxicologists no longer have to choose between optimum results and ease of use. Thermo Scientific™ ToxFinder™ software delivers highly accurate screening and semiquantitative analysis from either triple quadrupole or high-resolution, accurate-mass mass spectrometer data. Its experiment-specific workflows simplify operation by displaying only the features and parameters required for the experiment type selected. Data review is simple, yet powerful while the custom report designer provides WYSISYG customization
Screening and quantitation in a single workflow
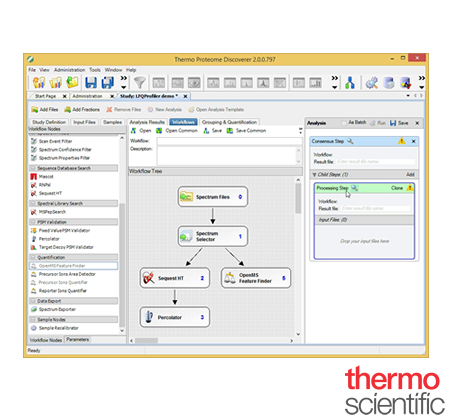
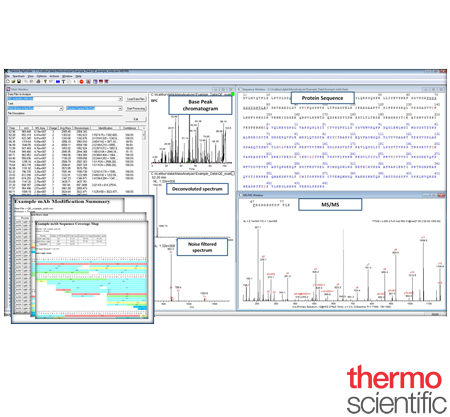
Proteome Discoverer™
Identify and quantify proteins in complex biological samples using Thermo Scientific™ Proteome Discoverer™ software. Proteome Discoverer software simplifies a wide range of proteomics workflows, from protein and peptide identification to PTM analysis to isobaric mass tagging and both SILAC and label-free quantitation. It supports multiple database search algorithms (SEQUEST, Z-Core, Mascot, and Byonic) and multiple dissociation techniques (CID, HCD, ETD, and EThcD) for more comprehensive analyses
Productive and Expandable
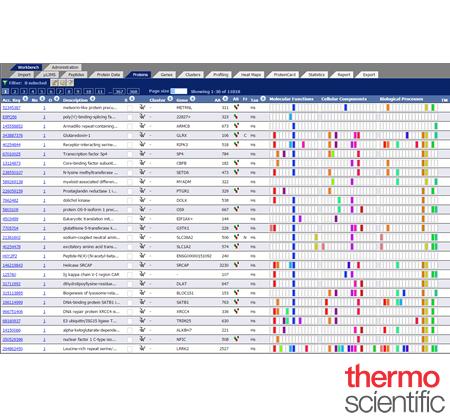
ProteinCenter™
Quickly extract meaningful biological information from multiple, complex data sets within minutes using Thermo Scientific™ ProteinCenter software, a web-based data tool for interpreting proteomics data. ProteinCenter software will find salient trends in data to facilitate biological insights
Accesss the Latest Data and >10 Million Non-redundant Proteins
PepFinder™
For biotherapeutic proteins to be effective, they must be produced in biologically active forms with proper folding and post-translation modifications (PTMs). Thermo Scientific™ PepFinder™ software provides accurate identification, in-depth characterization, and relative quantitation of biotherapeutic and other proteins from mass spectrometric data. It provides an automated workflows for glycopeptide identification, disulfide bond mapping, and quantification of PTMs. PepFinder software automates previously time-consuming manual processes, processing complex data and integrating the results into concise, informative reports
Easier Experimental Setup
Simglycan™
Speed the characterization of complex glycans using SimGlycan™ software from PREMIER Biosoft. SimGlycan software processes and interprets the multi-stage/sequential (MSn) data produced by Thermo Scientific™ Mass Spectrometers, a capability essential to determining glycan heterogeneity and isomeric forms. The software automatically matches experimental mass spectra against a comprehensive database and generates a scored list of candidate structures. More relevant information about the proposed structure, including the glycan class, pathway and enzyme, is provided via interactive links
Comprehensive Analyses
LipidSearch™
Lipidomics is crucial to understanding cellular physiology and pathology, and lipid profiling for disease phenotype analysis is a rapidly growing area in translational medical research. LC/MS is a powerful technique for the identification and quantitation of cellular lipids. The Thermo Scientific™ LipidSearch™ Software processes LC-MS data, including the high-resolution accurate-mass data generated by Thermo Scientific™ Orbitrap™-based mass spectrometers, to make provide accurate lipid identification. It automatically integrates complex data into reports and dramatically reduces data analysis time
Extensive Lipid Database for More Comprehensive Identification